
Overlapping regions with highly stable secondary structures may resist exo-nuclease activity. Because the In-Fusion reaction does not result in covalently joined DNA molecules, it is important to stick to recommended times for all steps in the procedure. The In-Fusion enzyme blend is proprietary to Takara Bio-sciences, entailing associated reagent costs.  Finally, the technique is relatively fast. The use of non-restriction and ligation cloning technology allows the direct fusion of desired fragments. The use of PCR means that reliance on conveniently located restriction enzymes is eliminated. In-Fusion Cloning: Pros & Cons In-Fusion ProsĪs one of several synthetic biology assembly techniques, In-Fusion is a sequence-independent and seamless cloning technique. This can lead to reduced reaction efficiency because the longer strands are unstable during DNA repair, and/or the longer single-stranded DNA may be subject to degradation. Adhere to the recommended fifteen-minute incubation.Įxcessive nuclease activity will create exposed single-stranded ends that are longer than desired. The goal is to expose the engineered overlaps between your fragments. The primary catalytic function of the In-Fusion reagent is its exonuclease activity. Multi-Fragment Insertion When incorporating multiple fragment inserts into one vector, start with a molar ratio of 2 molar units for each insert for each single molar unit of vector, i.e., 2:2:1 for two inserts into one vector. Smaller inserts may require even greater molar ratios. As your insert fragment decreases in length (150 base pairs to 500 base pairs), you may wish to increase the molar ratio to between 3:1 and 5:1 insert to vector. Single Fragment Insertion If your fragment and vector are similar in size, a molar ratio of 1:1 to 2:1 insert to vector is recommended. Takara Biosciences recommends the following molar ratios: Quantitation of your inserts is important but not essential for In-Fusion Cloning. In contrast, the In-Fusion exonuclease reaction is performed at 50☌, allowing some secondary structures to interfere with the exonuclease. Due to their length, many primer tails do not form stable secondary structures during PCR, where the reaction temperature ranges from 60☌ to 75☌. Multi-Fragment Insertions: Overlapping regions 20 base pairs or more increases cloning efficiency.Īs Primer tails increase in length, pay attention to the stability of the secondary structures that the tails can form. Single Fragment Insertion: Overlapping regions of 12-15 base pairs are sufficient. If you plan on using In-Fusion to clone various inserts into the same vector, you can design primer tails to be used on all pairs of primers and create one master vector preparation to use for all your cloning reactions. Vectors may be linearized using restriction enzyme digestion with either one or two enzymes selected to cut at the insertion point.Ĭomplementary ends in in-fusion cloning.
Finally, the technique is relatively fast. The use of non-restriction and ligation cloning technology allows the direct fusion of desired fragments. The use of PCR means that reliance on conveniently located restriction enzymes is eliminated. In-Fusion Cloning: Pros & Cons In-Fusion ProsĪs one of several synthetic biology assembly techniques, In-Fusion is a sequence-independent and seamless cloning technique. This can lead to reduced reaction efficiency because the longer strands are unstable during DNA repair, and/or the longer single-stranded DNA may be subject to degradation. Adhere to the recommended fifteen-minute incubation.Įxcessive nuclease activity will create exposed single-stranded ends that are longer than desired. The goal is to expose the engineered overlaps between your fragments. The primary catalytic function of the In-Fusion reagent is its exonuclease activity. Multi-Fragment Insertion When incorporating multiple fragment inserts into one vector, start with a molar ratio of 2 molar units for each insert for each single molar unit of vector, i.e., 2:2:1 for two inserts into one vector. Smaller inserts may require even greater molar ratios. As your insert fragment decreases in length (150 base pairs to 500 base pairs), you may wish to increase the molar ratio to between 3:1 and 5:1 insert to vector. Single Fragment Insertion If your fragment and vector are similar in size, a molar ratio of 1:1 to 2:1 insert to vector is recommended. Takara Biosciences recommends the following molar ratios: Quantitation of your inserts is important but not essential for In-Fusion Cloning. In contrast, the In-Fusion exonuclease reaction is performed at 50☌, allowing some secondary structures to interfere with the exonuclease. Due to their length, many primer tails do not form stable secondary structures during PCR, where the reaction temperature ranges from 60☌ to 75☌. Multi-Fragment Insertions: Overlapping regions 20 base pairs or more increases cloning efficiency.Īs Primer tails increase in length, pay attention to the stability of the secondary structures that the tails can form. Single Fragment Insertion: Overlapping regions of 12-15 base pairs are sufficient. If you plan on using In-Fusion to clone various inserts into the same vector, you can design primer tails to be used on all pairs of primers and create one master vector preparation to use for all your cloning reactions. Vectors may be linearized using restriction enzyme digestion with either one or two enzymes selected to cut at the insertion point.Ĭomplementary ends in in-fusion cloning. 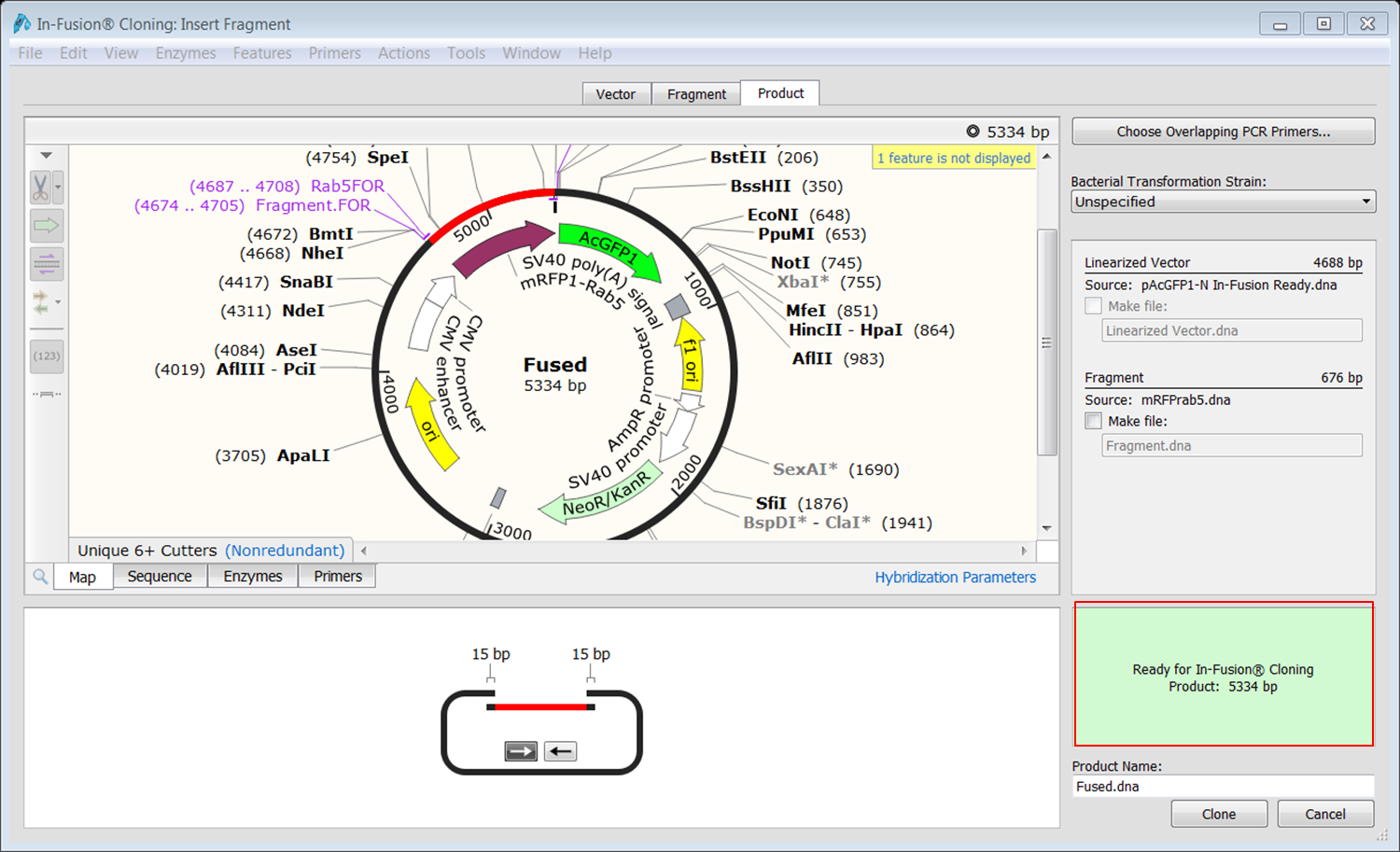
The two main methods of vector linearization are restriction enzyme digestion and inverse PCR. Any vector suitable for the downstream experimental use can be used in the In-Fusion reaction. The first step in In-Fusion cloning is to linearize your vector of interest at the insertion site.
Performing the In-Fusion reaction and validating the fusion points. Designing PCR primers to amplify your insert of interest and add the necessary tail for annealing to your vector. Linearizing your vector at the insertion point of interest. The three main steps when designing your In-Fusion experiment are: In-Fusion cloning allows you to add any insert into any vector at any site making it an extremely versatile cloning method. Designing an In-Fusion Cloning Experiment In-Fusion Cloning technique using a single insert.






 0 kommentar(er)
0 kommentar(er)
